Introduction
I often work in environments where I need to use Jupyter notebooks. I also prefer editing code in vim, keeping reusable logic in clean modules, and working primarily from the terminal. This post shows how I combine iron.nvim and Jupytext to work effectively with notebooks without leaving that environment.
Challenges with Notebooks
Notebooks are great for exploration, but there are some quirks when you try to integrate them into a more code-centric workflow:
- Execution order and state can be implicit rather than explicit
- Git diffs and review are harder than plain text
- The browser UI favors exploration over editing
This post isn’t intended as a critique of notebooks (there are many thoughtful takes on that already).
I firmly believe that notebooks have their advantages, and I do a lot of exploratory work in them. Many of the reasons I cited above can be fixed by working directly with a REPL.
The trouble I had was figuring out how to keep the fast, iterative feel of notebooks while still having a reproducible, script-based workflow.
Enter iron.nvim
At its core, iron.nvim is a vim plugin to send blocks of code to a REPL. Strictly it doesn’t even have to be for python, but that’s where I primarily use it. This gives me notebook-like iteration, but with explicit execution order and plain-text files.
The basic method is:
- Open a script in vim
- Highlight a block of text
- Send it to the REPL
- See outputs, etc., but also maintain a clear order of how and when code was run.
A simple example
For example, I setup a basic script to do some plotting:
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
x = np.linspace(0, 2 * np.pi, 10)
y = np.sin(x)
df = pd.DataFrame({'x': x, 'y': y})
fig, ax = plt.subplots()
sns.lineplot(
data=df,
x='x',
y='y',
ax=ax
)
fig.show()
Here’s the same workflow using iron.nvim.
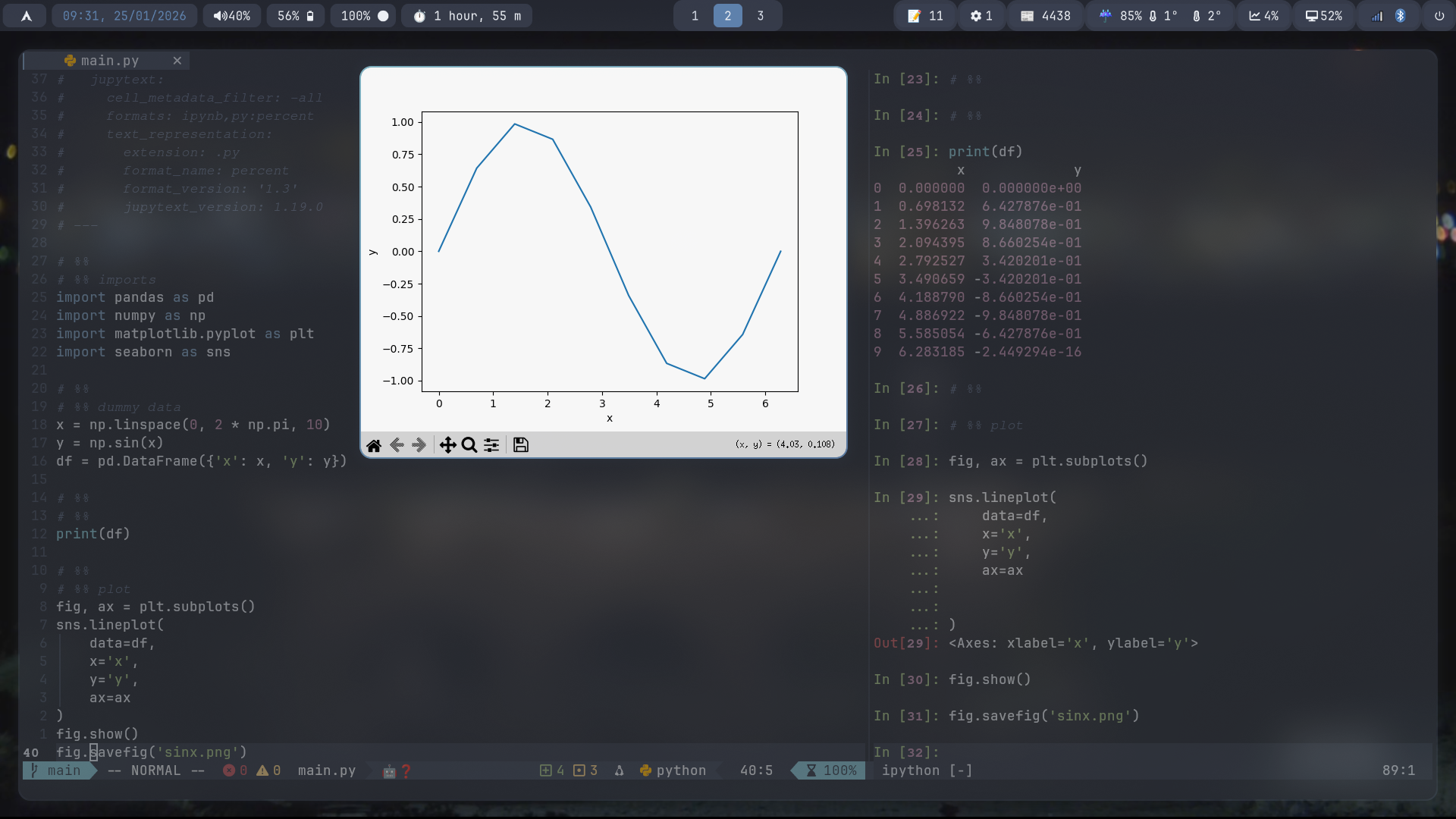
This means plots aren’t inline by default. Instead, I treat them like regular artifacts, saved to disk and viewed explicitly.
Aside: viewing plots
There are a few options to do this:
- opening figures in external windows
- using vim’s ability to run shell commands e.g.
! sxiv my_plot.png
Or what I do: switching to a tmux pane where plots are saved and opening from the terminal (and even using kitty’s graphics protocol1, probably for another post)
How to convert to a jupyter notebook and back
Jupytext is a fantastic tool, which allows you to sync a python file with a jupyter notebook and seamlessly convert between the two. The core idea is that the notebook and the script are two views of the same underlying content.
The basic syntax is:
# set formats
jupytext --set-formats ipynb,py main.py
# convert between notebook/script
jupytext --to py notebook.ipynb
jupytext --to ipynb notebook.py
Once formats are linked, ‘cells’ can be created with the ‘# %%’ syntax, with special ‘[markdown]’ formatters for markdown cells. E.g.
# %%
import numpy as np
# %% [markdown]
# ## Plotting
For example, the notebook corresponding to my example:
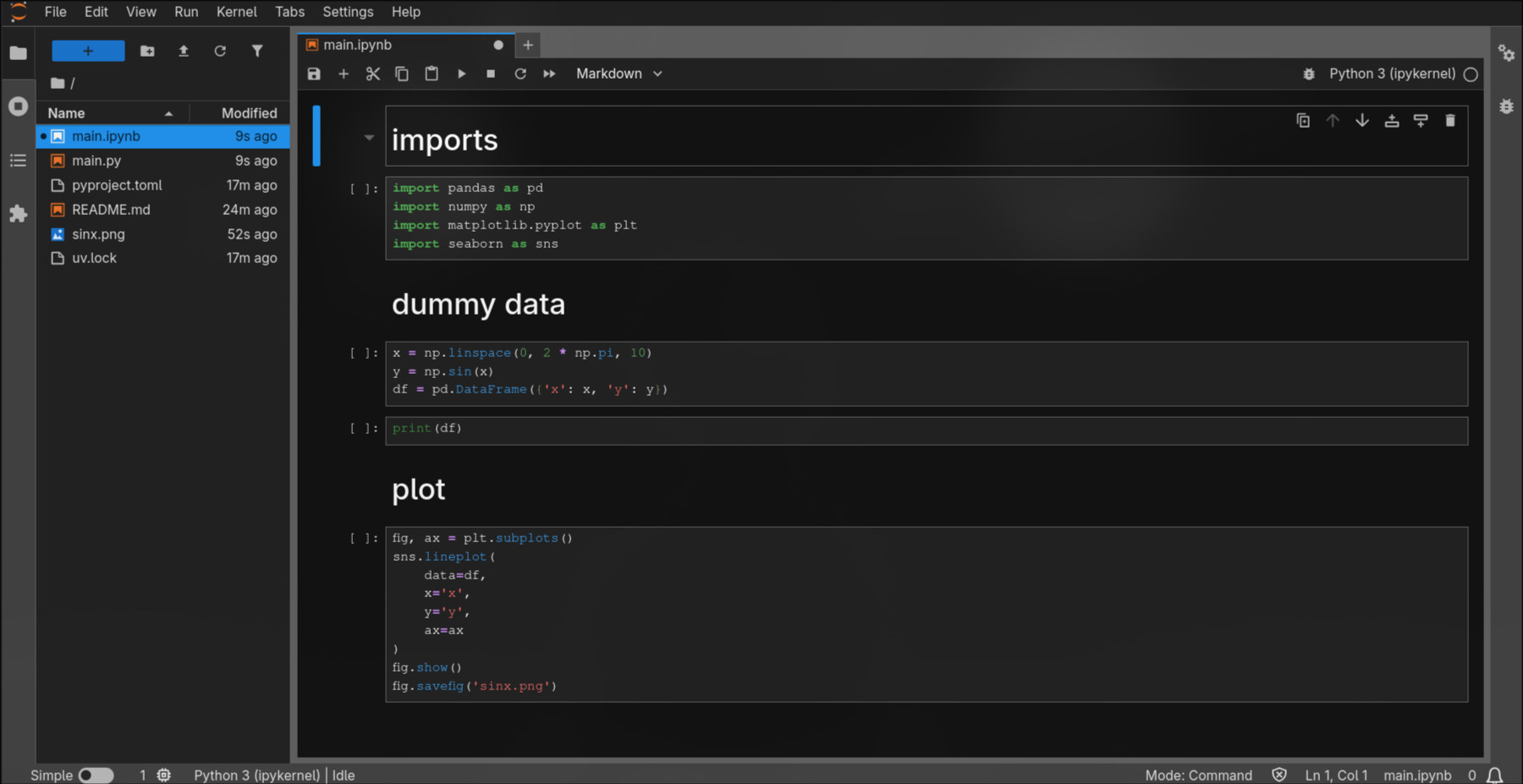
Collaboration
To me, jupytext is essential here. It allows me to convert between notebooks and scripts easily, and most importantly allows me to share work.
The key is flexibility: meet your collaborators where they are whilst keeping your workflow efficient.
My collaborators see a normal .ipynb file and don’t need to change how they work.
I get clean diffs, reproducible execution, and vim-native editing. Everyone is happy.
Summary
This setup won’t replace notebooks for exploration — and it’s not meant to. It’s simply a way to work comfortably in vim without making that everyone else’s problem.
Example repository
The examples showcased in this post can be found at this repository: https://github.com/sdysch/iron_nvim_jupytext_test/.
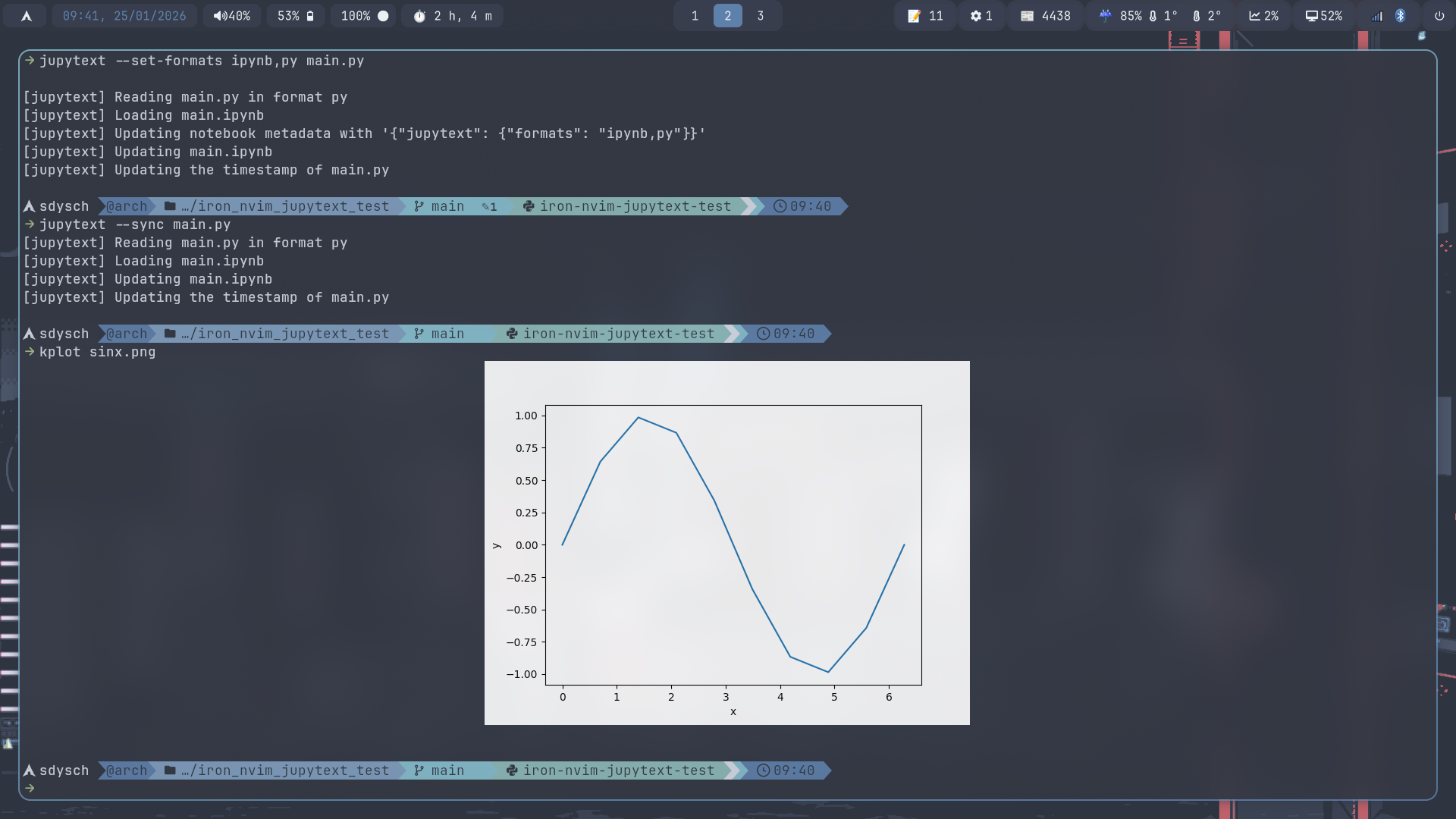 I have
I have bash
alias kplot="kitty +kitten icat"